The ABCs of Influenza: Evolution, Transmission, and Pandemic Potential
The ABCs of Influenza Viruses
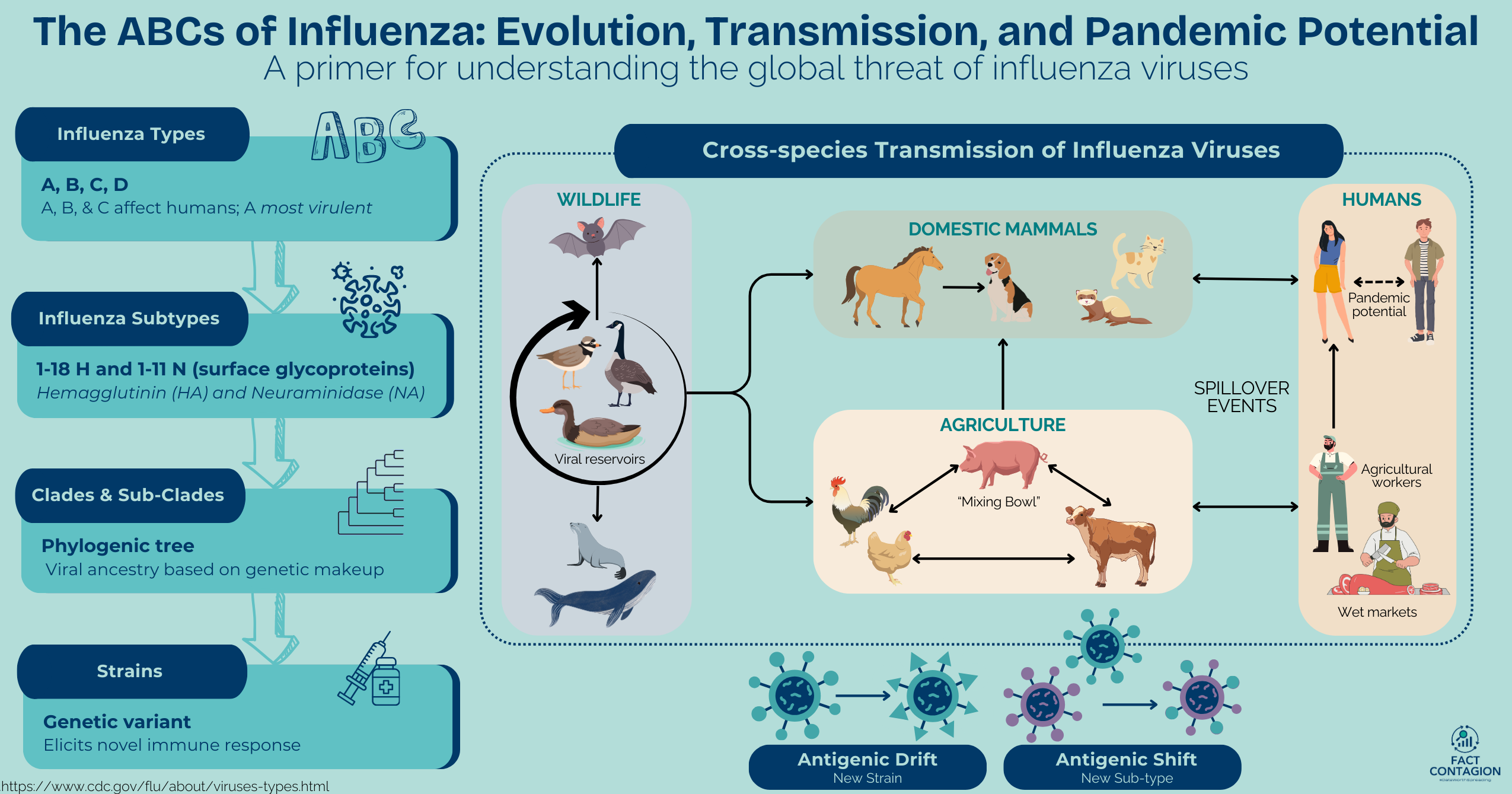
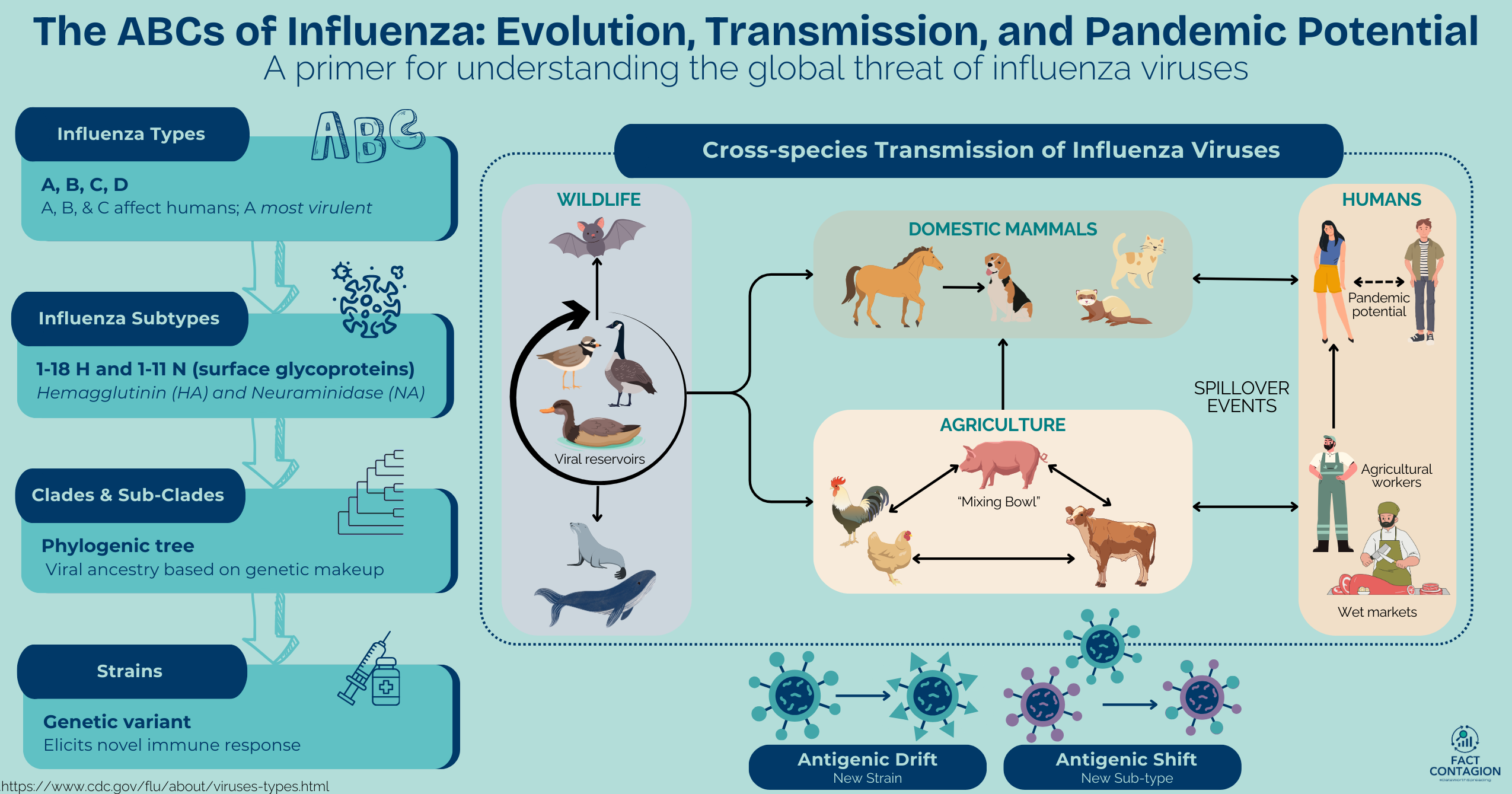
There are four species, or types, of influenza viruses (A, B, C, and D). Humans can be infected with types A, B, and C, but influenza A is the most virulent (severe). Wild aquatic birds are the natural reservoirs for most influenza A viruses, but through various modes of transmission and evolution, influenza A viruses can also infect domestic birds, and a wide range of mammals, including humans, cattle, pigs, horses, dogs, cats, and even marine mammals!
Influenza A viruses are further classified by subtype (serotype), which is based on two main surface glycoproteins (GPs): hemagglutinin (HA) and neuraminidase (NA). 18 HA and 11 NA GPs are recognized. They help cells communicate, allow the immune system to recognize host vs foreign, and can form protective barriers like mucus. Viruses often use GPs to attach to and enter cells (like COVID’s Spike protein). Subtypes of influenza A currently circulating among people worldwide are H1N1 and H3N2 viruses. Influenza A & B viruses are further classified into different HA genetic clades and sub-clades. These are visually displayed via phylogenetic trees (akin to a family’s genealogy tree) that group viruses with a similar genetic makeup and common viral ancestor. Influenza A subtypes are further classified by strain.
What makes influenza so unique?
Influenza is an RNA virus. In general, RNA viruses are more unstable than DNA viruses, because of the error-prone RNA synthesis. Their polymerases that build the RNA strands are pretty bad at proof-reading their work.
Stick with me here through more science jargon… What makes influenza even more unique is its segmented ribonucleoprotein (RNP). RNP is a bundle of RNA and protein that join together to perform important tasks in cells, like a toolkit of sorts. Influenza A contains 8 gene segments: PB1, PB2, PA, NP, M, NS, HA, and NA. When a virus infects a living cell, its genome is transported into the cell’s nucleus where the 8 segments undergo transcription (copying DNA’s recipe to form mRNA) and translation (making the meal from mRNA’s recipe by assembling amino acids into proteins). The viral protein segments then exit the nucleus and reassemble near the cell membrane.
Influenza A's pathogenesis (how it affects the body) relies on several virus and host characteristics, with a key virulence (severity) factor being the HA glycoprotein. HA facilitates virus binding to cell receptors and introduction of viral RNP into the cell. HA also induces the host’s antibody response to prevent infection.
Influenza’s Evolutionary Playbook: It’s About Drift and Shift
Influenza viruses evolve by distinctive mechanisms.
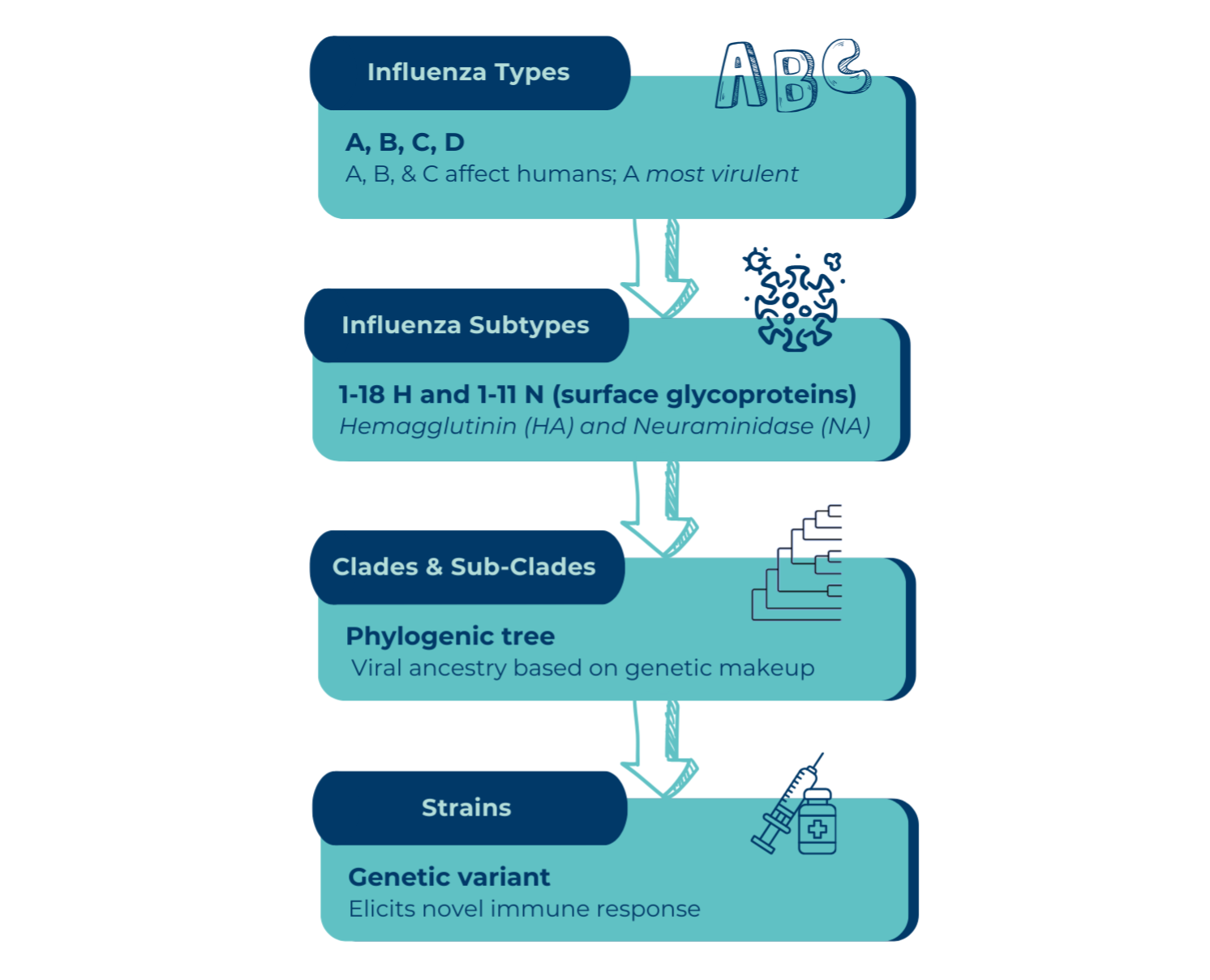
Antigenic drift creates new strains within an influenza subtype due to minor, unpredictable point mutations in the genes that produce the HA and NA surface GPs. This continual "drifting" is why seasonal flu vaccines must be updated annually. New strains may not be recognized by our immune systems (from previous infections or vaccines); therefore, we can be infected with multiple strains of the same influenza subtype. Experts meet annually to try to predict which strains will cause the majority of seasonal influenza infections the following year. Two influenza A viruses and 1 B virus make up the seasonal vaccine. And yes, it sometimes is shot in the dark. Some years they're right, some years they are not.
The other type of evolution is called antigenic shift, which can occur by two processes:
1) An abrupt, major change of an influenza virus can lead to direct species-to-species transmission (such as wild bird-to-pig transmission, or cattle-to-human transmission). This is a notably rare event for any influenza virus.
2) Two or more influenza viruses can mix in an intermediate host to create a new influenza subtype (new HA or NA). For example, a human virus and an avian virus mixing in a pig. Because influenza’s genome is segmented, if two influenza viruses infect the same cell, then each of their 8 segments can intermix during the assembly stage and produce a new progeny virus with components of both parent viruses. This exchange of genetic material between multiple influenza viruses is called reassortment. Think of it like shuffling a deck of cards.
In both instances of antigenic shift, people and animals will have no immunity to this new virus. The virus would be able to spread rapidly through a community and potentially lead to a pandemic.
Cross-species transmission and the swine "mixing bowl" theory
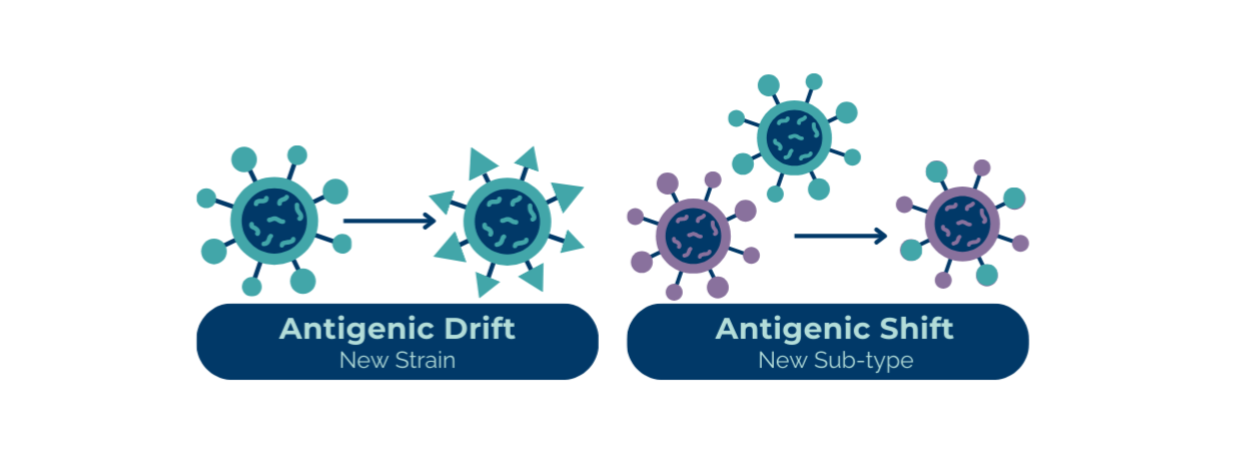
It has been suggested that some of influenza's host specificity relies on specific cell receptors in the host's respiratory tract. The HA and NA surface glycoproteins of avian and cattle influenza viruses bind to different sialic acid receptors in the host’s respiratory tract (eg, α2,3) than swine and human influenza viruses (eg, α2,6). Pigs' respiratory tracts express both α2,3 and α2,6 linked receptors.
Therefore, when considering cross-species transmission, humans are more readily infected with swine influenza viruses than avian and cattle influenza viruses because the swine viruses also bind to the α2,3-linked receptor. Human infections with avian or cattle influenza (and avian infections with swine and human influenzas) do still occur but are less common. Because swine have cell receptors appropriate for both avian and mammalian HA subtypes, they effectively serve as a “mixing bowl” for the reassortment of viruses from different species. Avian and human influenza viruses that co-infect the same cell in one pig could lead to reassortment of viral genes and potentially novel progeny viruses.
Until recently, the swine “mixing bowl” theory was the central theory regarding the emergence of pandemic strains. However, the increasing number of direct avian-to-human influenza infections reported have shaken that popular theory. While it is still a valid means in which reassortment can occur, recent events illustrate that influenza viruses have other means in which to cross the species barrier. Fortunately, thus far, human infections have been relatively few and incidental compared to larger scale outbreaks in wild birds and domestic poultry and cattle.
Pandemic Potential: Why Influenza Is a Global Threat
Influenza pandemics occur when:
1) A novel virus subtype emerges that
2) Causes serious illness in humans who have little or no immunity against the virus and
3) Spreads easily from person to person.
Often the human immune system is naive and susceptible because these novel viruses present with antigens of animal virus origin. Whereas the pandemics of 1918, 1957, and 1968 had genetic components from avian viruses, pandemic viruses may arise from any number of animal influenza viruses, such as the case with the most recent pandemic H1N1 (2009) virus. This novel H1N1 pandemic virus had a lineage of human, swine, and avian origins.
Up Next
With this primer of the basics, next we’ll dive into the latest twist: highly pathogenic H5N1 avian influenza’s surprising impact on dairy cattle in the U.S.
Reference: https://www.cdc.gov/flu/about/viruses-types.html

