Bird Flu Unmasked: Tracing the Roots of HPAI H5N1
First off, if you haven’t already, check out my primer on influenza virus biology and what makes flu a global threat:
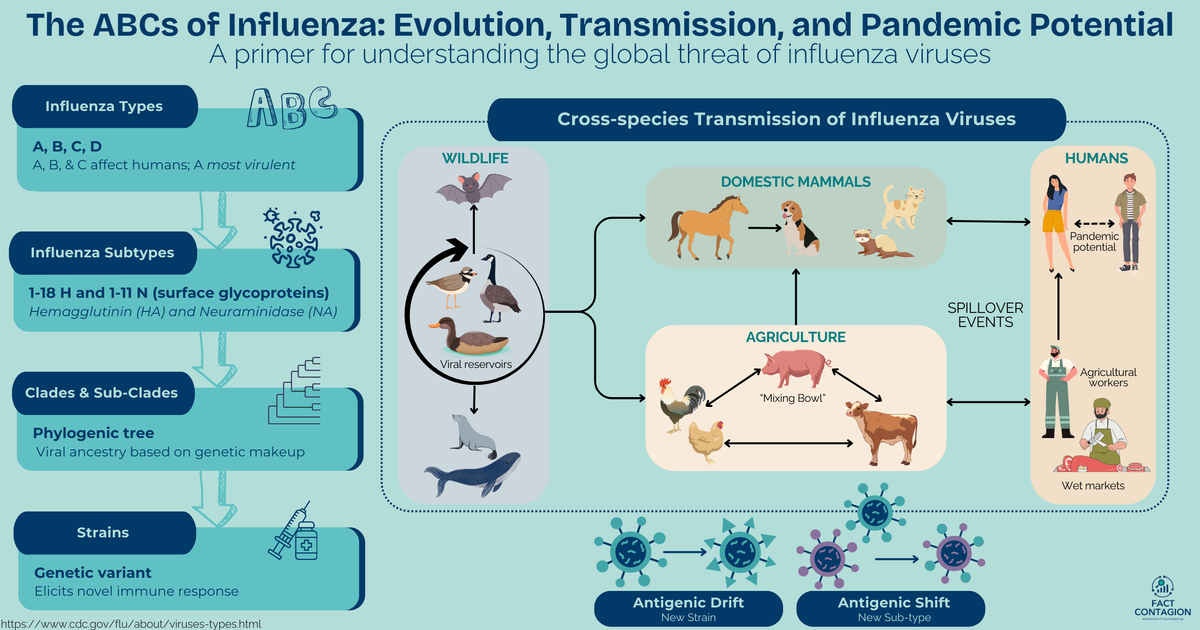
The Perfect Storm: Factors Driving the Spread of Avian Influenza
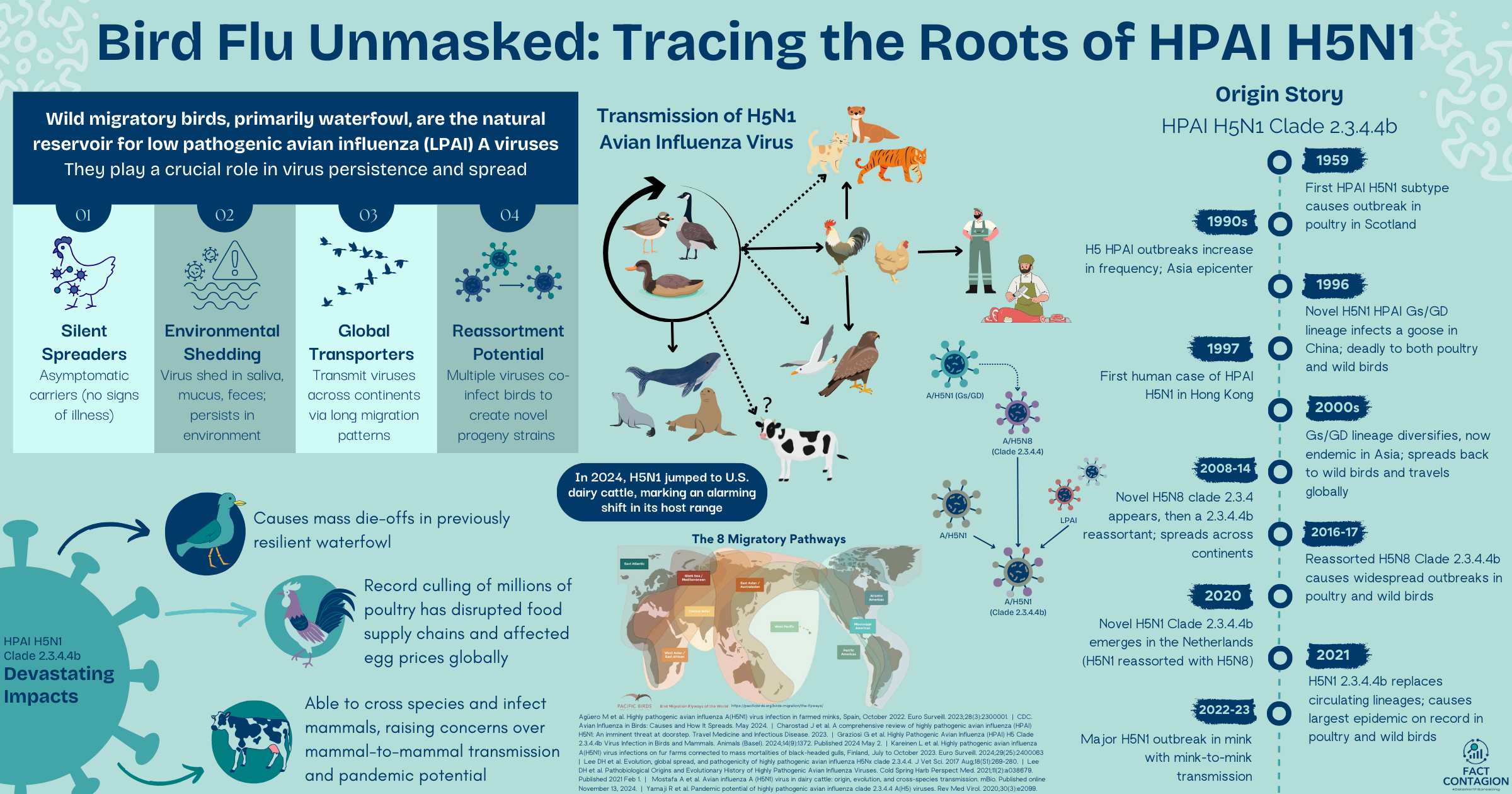
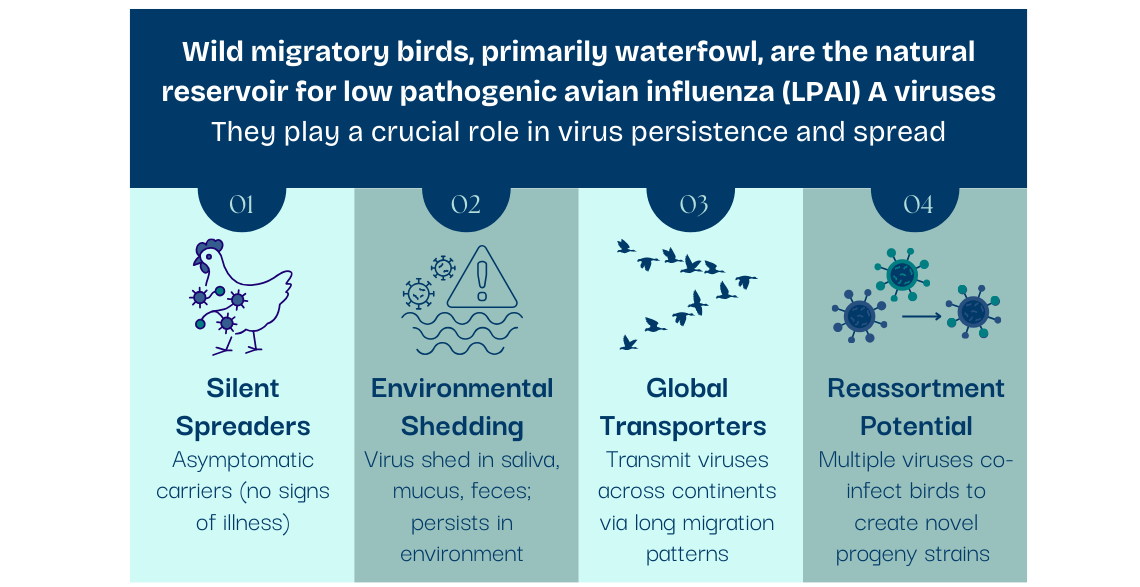
Wild migratory birds, primarily waterfowl like ducks, geese, swans, gulls, and terns, are the natural reservoir host for low pathogenic avian influenza (LPAI) A viruses (“bird flu”). They harbor almost all influenza subtypes: 17 of the 19 known hemagglutinin (HA) and 9 of the 11 known neuraminidase (NA) subtypes have been found in wild waterfowl.
Wild aquatic birds play a crucial role in the persistence and spread of influenza.
Asymptomatic Carriers: Wild aquatic birds can be infected with avian influenza viruses without showing any signs of illness, allowing the virus to spread within and across flocks undetected.
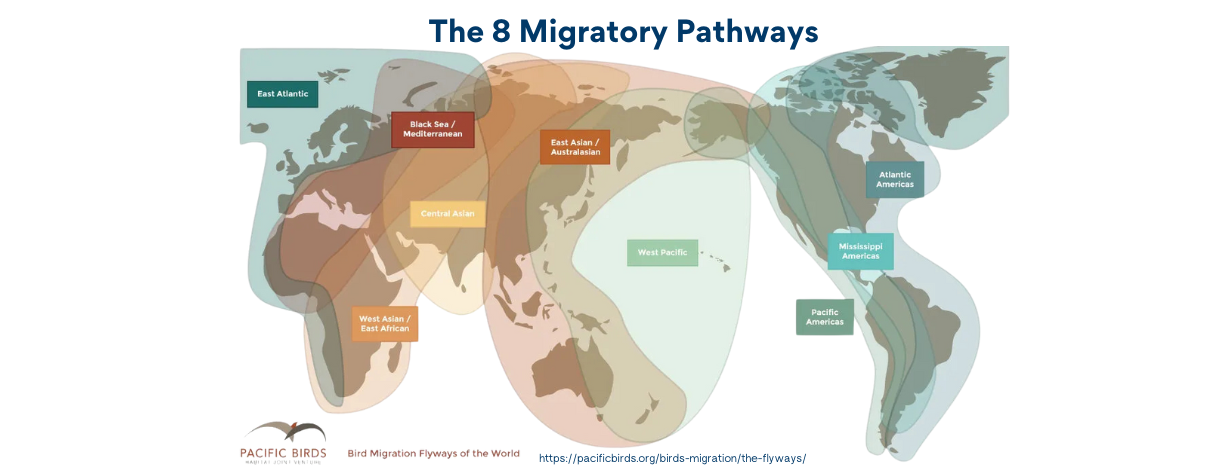
Migration Patterns: These birds embark on long-distance migrations, transmitting viruses across continents and introducing strains into new regions.
Environmental Shedding: Infected birds shed virus in their saliva, mucus, and feces. Virus can persist in water and soil and spread to susceptible birds.
Reassortment Potential: Multiple avian influenza virus subtypes can co-infect a bird, allowing for the viruses to exchange genetic segments and create novel progeny virus strains with altered characteristics, such as increased virulence or transmissibility.
These features create a perfect storm, where wild aquatic birds supply a continual source of LPAI viruses that not only fuels outbreaks in domestic poultry but also creates opportunities for viruses to adapt and gain the ability to spillover into other animal species, including humans.
HPAI H5N1’s Origin Story
Avian influenza A viruses are classified as either "low pathogenic" or "highly pathogenic" based on key genetic traits and the severity of disease they cause in poultry. Highly pathogenic avian influenza (HPAI) viruses evolve from low pathogenic avian influenza (LPAI) viruses through genetic changes in the hemagglutinin (HA) surface glycoprotein, which plays a critical role in infection.
The history of HPAI dates back to the 1880s! It was referred to as the “fowl plague”. After first appearing in chickens in northern Italy in 1878, geographically dispersed outbreaks occurred in Europe, Asia, Africa, and the Americas, caused by H7N7 and N7N1 virus strains.
It was 1959 when the first H5N1 subtype of HPAI evolved from a LPAI virus in wild birds and appeared in chickens in Scotland. This evolution marked a turning point for the avian influenza virus.
For the next 30 years, sporadic, localized outbreaks in poultry of HPAI caused by H5, as well as H7, subtypes occurred across Asia. The virus was effectively contained through culling and other biosecurity measures. These HPAI H5 and H7 viruses did not recirculate in wild birds.
Then in the early 1990s, H5 HPAI outbreaks in domestic poultry began increasing in frequency across Asia. Asia was becoming an epicenter for HPAI outbreaks, attributable to their intensive farming practices and live bird (wet) markets that provided ideal conditions for its evolution and spread.
A Proverbial Game Changer
It’s now 1996. In southern China. A goose in Guangdong Province becomes the host to a novel Gs/GD lineage of H5N1 HPAI (A/goose/Guangdong/1/1996 (Gs/GD)). This strain was a proverbial game changer. Not only had it developed genetic mutations making it lethal to both domestic poultry and wild birds, but it set the stage for global concerns about its potential to spill over into other species, including humans.
Shortly later in 1997, HPAI H5N1 spilled over into humans. It was an outbreak in Hong Kong, involving 18 people who had close contact with wet markets; 6 of whom died. This was a stark warning of the virus’s zoonotic potential and pandemic threat.
Through the 2000s and into the 2010s, HPAI H5N1 continued to evolve through genetic mutations and reassortment with other influenza A viruses. The Gs/GD lineage adapted into 10 genetically distinct virus clades (0–9) and multiple subclades. After becoming endemic in domestic poultry in Asia, the Eurasian H5 HPAI viruses from the Gs/GD phylogenetic lineage found their way back into wild migratory birds, allowing it to escape beyond Asia, reaching Europe, the Middle East, Africa, and the Americas. The virus adapted to various hosts and environments. Control efforts became more complicated. Sporadic human cases continued to be reported, chiefly among individuals with close contact with infected poultry. Human-to-human transmission was limited, but the high mortality rate was ringing alarm bells.
The Rise of Clade 2.3.4.4b: The Next Chapter for H5N1
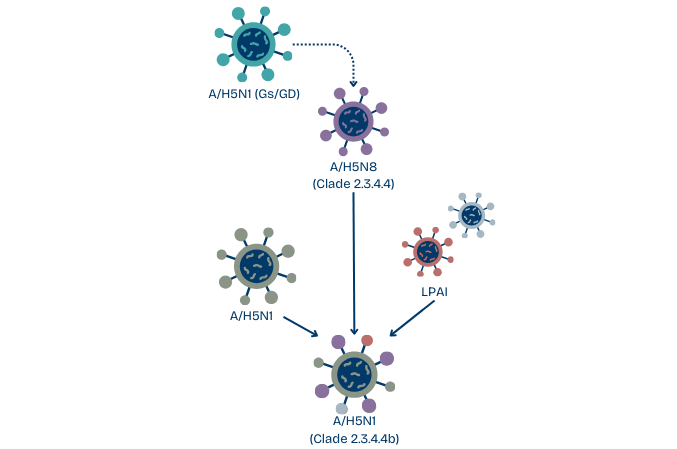
Since 2008, subtypes H5N2, H5N5, and H5N8 of the Gs/GD lineage H5 clade 2.3.4 began appearing in domestic poultry (mainly wet markets) in China.
In 2010, a H5N8 subtype of clade 2.3.4.4 was isolated from a Chinese wet market. It went on to cause several outbreaks in poultry and wild birds in South Korea. Since then, the H5N8 clade 2.3.4.4 experienced further reassortment events with H5N1 and LPAI viruses.
In 2013-14, clade 2.3.4.4 split in four different genetic groups (Groups A-D). H5N8 viruses belonging to Group B (Clade 2.3.4.4b) were identified in China in 2013-14 and Korea in 2014.
In the fall of 2016, novel H5N8 clade 2.3.4.4b reassortant viruses containing 5 LPAI gene segments were discovered in wild birds found dead in China and Siberia. The clade further reassorted with Eurasian LPAI viruses and spread from Siberia into Europe, Africa, the Middle East, and Asia via the southward migration of waterfowl, where over a thousand poultry outbreaks and nearly 1600 wild bird die-offs were reported from 2016-17.
HPAI H5Nx Clade 2.3.4.4b viruses continued to diversify across 4 additional genetic groups (E-H) from 2018-2020, with sporadic outbreaks occurring in Eastern Europe.
Emergence of H5N1 in Clade 2.3.4.4b
Then, in October 2020, a paradigm-shifting novel variant of H5N1 belonging to clade 2.3.4.4b emerged in the Netherlands through multiple reassortment events between H5N8 Clade 2.3.4.4b virus, an H5N1 virus, and several other LPAI.
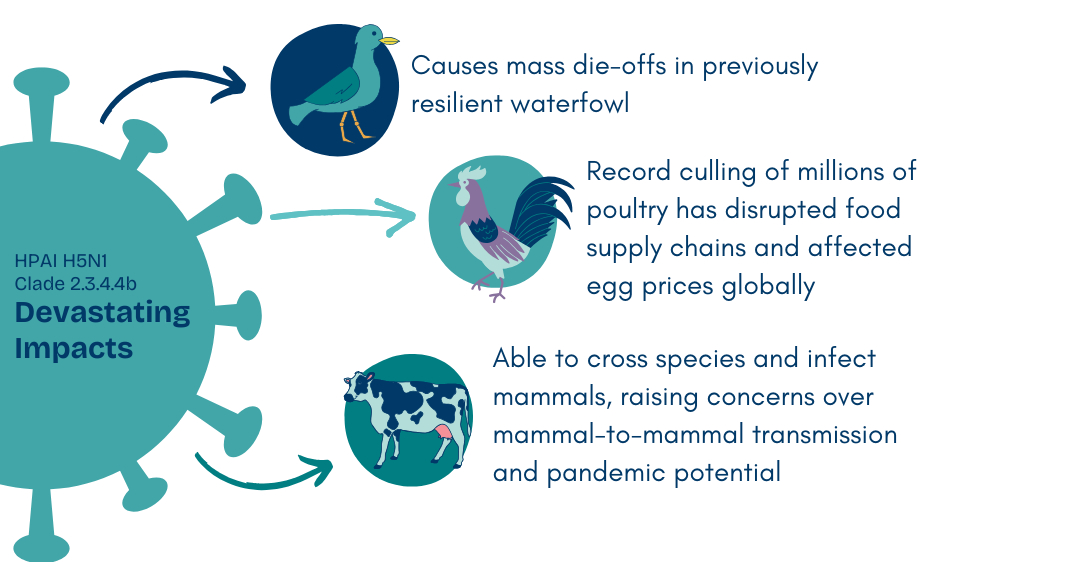
This H5N1 virus exhibited unprecedented adaptability and virulence. More than 19 different H5N1 clade 2.3.4.4b genotypes emerged from reassortment events. By 2021, this clade effectively replaced all other circulating avian influenza lineages and caused widespread outbreaks across Asia, Europe, and the Americas via migratory birds. This peaked in 2021-22, with a record number of outbreaks in poultry and cases among wild birds. As the largest HPAI epidemic to date, more than 2000 outbreaks across 37 European countries were reported and over 40 million birds were culled.
You may remember the effect this had on egg prices in the US. By January 2023, the average cost for a dozen eggs in the US surged to $4.82. The the culling of millions of egg-laying hens reduced supply and drove up costs. (Spoiler: there are signs prices are again going up, thanks to the latest wave of bird flu and holiday baking demands.)
What set H5N1 in Clade 2.3.4.4b further apart was its devastating impact on wild waterfowl. While earlier H5 HPAI clades often caused mild or asymptomatic infections in its natural host, this new variant was triggering significant morbidity and mass die-offs in wild waterfowl. This increased pathogenicity emphasized the critical need for international vigilance and coordinated strategies for surveillance and control.
H5N1 Clade 2.3.4.4b’s Jump Into Mammals
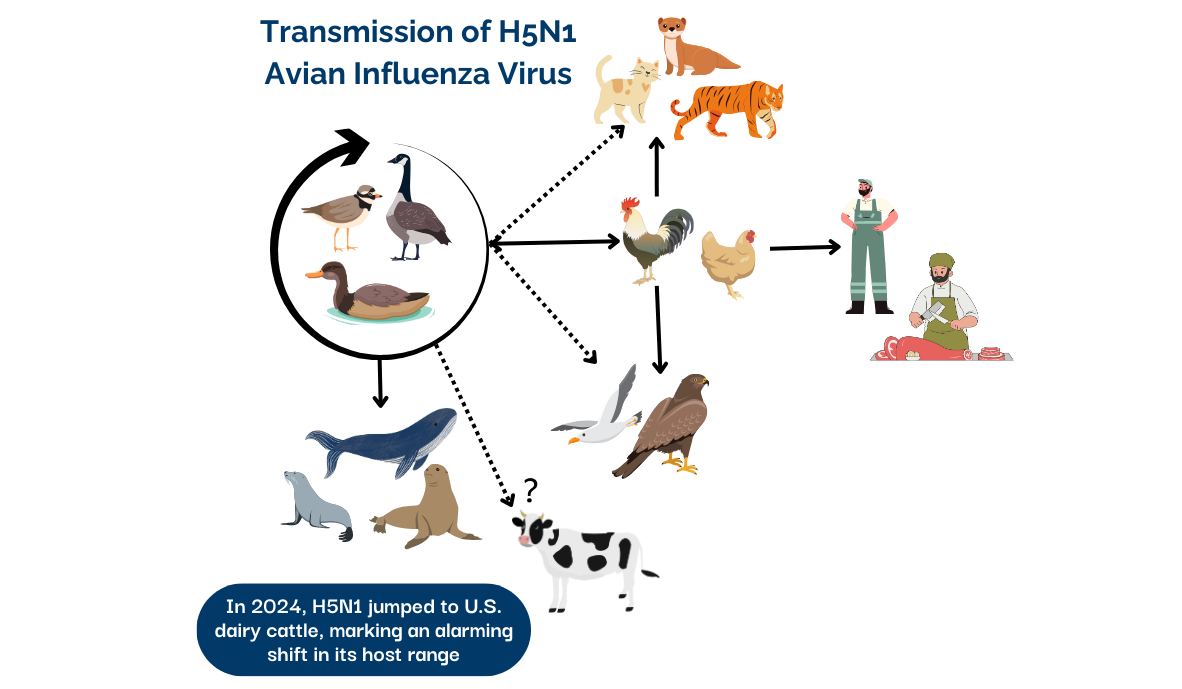
H5N1 Clade 2.3.4.4b soon began infecting mammals, including cats, tigers, seals, and sea lions. Concerns mounted over its ability to cross species barriers. The list of affected species continued to grow, as the virus was adapting and expanding its host range.
In late 2022, the first major outbreak in mammals was reported at a mink farm in Spain that then spread across numerous farms in Finland through 2023. And yet another unprecedented revelation: high viral loads detected in swab samples, along with rapid spread across farms, suggested efficient mink-to-mink transmission. In South America, mass die-offs of sea lions provided further evidence of H5N1’s ability to spread between mammals, signaling another troubling shift in its transmission dynamics. Mammal-to-mammal transmission is a critical development that increases the risk of adaptation for human-to-human transmission.
A Worrisome New Development: H5N1 Clade 2.3.4.4b Emerges in American Dairy Herds
In 2024, H5N1’s host range again expanded to include ruminants, particularly dairy cattle in the US. Stay tuned for the next installment that will cover the new twist in H5N1 Clade 2.3.4.4b's evolution: its leap into US dairy cows.
References and Further Reading
Agüero M, Monne I, Sánchez A, et al. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Euro Surveill. 2023;28(3):2300001. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2023.28.3.2300001
CDC. Avian Influenza in Birds: Causes and How It Spreads. May 2024. https://www.cdc.gov/bird-flu/virus-transmission/avian-in-birds.html
Charostad J, Rukerd M, Mahmoudvand S, Bashash D, Hashemi S, Nakhaie M, Zandi K. A comprehensive review of highly pathogenic avian influenza (HPAI) H5N1: An imminent threat at doorstep. Travel Medicine and Infectious Disease. 2023. https://doi.org/10.1016/j.tmaid.2023.102638
Durbin DA, Funk J, Vancleave M. AP News. Eggs are getting pricier ahead of the holiday baking season. Nov 26, 2024 https://apnews.com/article/eggs-prices-bird-flu-7f2063d64dd80ec4ed0aa74e562f19d7
Graziosi G, Lupini C, Catelli E, Carnaccini S. Highly Pathogenic Avian Influenza (HPAI) H5 Clade 2.3.4.4b Virus Infection in Birds and Mammals. Animals (Basel). 2024;14(9):1372. Published 2024 May 2. https://www.mdpi.com/2076-2615/14/9/1372
Kareinen L, Tammiranta N, Kauppinen A, et al. Highly pathogenic avian influenza A(H5N1) virus infections on fur farms connected to mass mortalities of black-headed gulls, Finland, July to October 2023. Euro Surveill. 2024;29(25):2400063 https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2024.29.25.2400063
Lee DH, Bertran K, Kwon JH, Swayne DE. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci. 2017 Aug;18(S1):269-280. https://vetsci.org/DOIx.php?id=10.4142/jvs.2017.18.S1.269
Lee DH, Criado MF, Swayne DE. Pathobiological Origins and Evolutionary History of Highly Pathogenic Avian Influenza Viruses. Cold Spring Harb Perspect Med. 2021;11(2):a038679. Published 2021 Feb 1. https://perspectivesinmedicine.cshlp.org/content/11/2/a038679.long
Mostafa A, Naguib MM, Nogales A, et al. Avian influenza A (H5N1) virus in dairy cattle: origin, evolution, and cross-species transmission. mBio. Published online November 13, 2024. https://journals.asm.org/doi/10.1128/mbio.02542-24
Yamaji R, Saad MD, Davis CT, et al. Pandemic potential of highly pathogenic avian influenza clade 2.3.4.4 A(H5) viruses. Rev Med Virol. 2020;30(3):e2099. https://onlinelibrary.wiley.com/doi/10.1002/rmv.2099


